作者简介:
梁骏(1981-),男,广东梅州人,主治医师,医学硕士,研究方向为老年危重患者的综合救治,重症患者消化系统疾病救治与机制研究。
目的 免疫检查点基因是调控结肠癌患者免疫应答的关键机制,并对结肠癌患者的预后、治疗具有潜在的临床应用价值,然而目前尚无基于免疫检查点基因的预后模型来预测结肠癌患者的预后和免疫状态。方法 从肿瘤基因组图谱(The Cancer Genome Atlas,TCGA)和高通量基因表达(Gene Expression Omnibus,GEO)公共数据库获取并整理结肠癌患者的基因表达谱数据及其对应的临床信息,鉴定差异表达的免疫检查点基因。采用最小绝对收缩和选择算子(least absolute shrinkage and selection operator,LASSO)Cox回归分析构建免疫检查点基因相关的TCGA结肠癌患者预后模型。应用GEO结肠癌患者队列(GSE143985)进行验证。Kaplan-Meier曲线和受试者操作特征(receiver operating characteristic,ROC)曲线用于评估模型预测准确性。Spearman相关性分析观察预后模型与免疫细胞浸润情况。单因素和多因素回归分析免疫检查点风险基因及临床指标,并基于多因素回归结果构建临床列线图。结果 在正常和结肠癌组织中共筛选出29个差异表达的免疫检查点相关基因,LASSO回归分析后构建了基于14个免疫检查点相关基因的预后模型。根据中位风险评分将TCGA结肠癌患者分为高、低危险组,Kaplan-Meier生存分析显示,相较于低危组,高危组患者的生存较差。ROC分析则显示TCGA训练队列的1年、3年预测曲线下面积(area under the curve,AUC)值均大于0.7,而GEO验证队列的1年、3年预测AUC值均大于0.8,显示了模型较高的准确性和稳定性。免疫细胞浸润分析显示风险评分与CD4+ T细胞和CD8+ T细胞有显著相关性。此外,基于免疫检查点风险基因及临床指标构建的列线图C-指数为0.767,显示了较高的临床应用价值。结论 我们构建了一种新的免疫检查点基因相关的结肠癌预后模型,可用于结肠癌的预后及免疫状态预测,为结肠癌患者的临床预后提供一定的指导依据。
Objective Immune checkpoint gene was a key mechanism regulating the immune response in patients with colon cancer and had potential clinical application value for the prognosis and treatment of patients with colon cancer. However, there was no prognostic model based on the immune checkpoint genes to predict colon cancer patients' prognosis and immune status.Methods The gene expression profile data and corresponding clinical information of colon cancer patients were collected from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) public databases and the differentially expressed immune checkpoint genes were identified. The least absolute contraction and selection operator (LASSO) Cox regression analysis was used to construct an immune checkpoint gene-related prognostic model of TCGA patients with colon cancer. The GEO colon cancer patient cohort (GSE143985) was used for validation. Kaplan-Meier curve and receiver operating characteristic (ROC) curve were used to evaluate the prediction accuracy of the model. Spearman correlation analysis was used to observe the prognosis model and immune cell infiltration. The immune checkpoint risk genes and clinical indicators were analyzed by univariate and multifactorial regression, and a clinical histogram was constructed based on the multivariate regression results.Results A total of 29 differentially expressed immune checkpoint-related genes were screened in normal and colon cancer tissues. After LASSO regression analysis, a prognostic model based on 14 immune checkpoint-related genes was constructed. The TCGA colon cancer patients were divided into high-risk and low-risk groups based on the median risk score. Kaplan-Meier survival analysis showed that patients in the high-risk group had poorer survival than those in the low-risk group. ROC analysis showed that the 1-year forecast area under the curve (AUC) values of the TCGA training cohort and the 3-year forecast AUC values were all greater than 0.7, while the 1-year forecast AUC values of the GEO verification cohort were all greater than 0.8, indicating the high accuracy and stability of the model. Immune cell infiltration analysis showed a significant correlation between risk scores and CD4+ T cells and CD8+ T cells. In addition, the C-index of the graph constructed based on immune checkpoint risk genes and clinical indicators was 0.767, indicating a high clinical application value.Conclusions We construct a new immune checkpoint gene-related prognostic model for colon cancer, which can be used to predict the prognosis and immune status of colon cancer patients and provide a certain guidance basis for the clinical prognosis of colon cancer patients.
结肠癌是一种最常见的消化系统恶性肿瘤, 与肺癌、前列腺癌和乳腺癌一起被认为是癌症四大杀手之一[1], 严重威胁人类的健康。手术以及化疗是目前治疗结肠癌的主要方法, 但晚期结肠癌患者的治疗疗效不高[2]。目前, 分子靶向治疗是治疗多种癌症的希望之一, 但由于缺乏完整的的分子机制了解, 临床治疗结肠癌的有效靶向治疗少于其他癌症。因此, 了解导致结肠癌发展的生物学途径, 并基于此开发新的结肠癌患者预后分层将对改善结肠癌预后以及制定合适的治疗策略至关重要。
近年来, 免疫检查点抑制剂治疗的发现促进了癌症中的精准治疗的发展。到目前为止, 多种免疫检查点靶向抑制剂已经被批准上市并用于治疗结肠癌, 其中包括程序性细胞死亡蛋白1(programmed cell death 1, PD-1)抑制剂帕博利珠单抗, 程序性细胞死亡配体1(programmed cell death ligand-1, PD-L1)抑制剂阿替利珠单抗, 细胞毒性T淋巴细胞相关抗原4(cytotoxic T lymphocyte-associated antigen-4, CTLA-4)抑制剂伊匹木单抗[3]。最近一项临床试验还评估了免疫检查点抑制剂联合瑞戈非尼治疗部分晚期或转移性微卫星稳定结肠癌患者的疗效, 并证明该治疗方案在总体上对难治性结肠癌患者有显著的无进展生存期改善[4]。此外, 有研究表明PD-L1在结肠癌中的表达显著升高, 其可以通过促进免疫抑制环境, 并允许免疫逃逸, 从而促进肿瘤生长[5]。还有一些研究指出, 免疫检查点相关的受体和配体, 如LAG3、CD80均参与了结肠癌的进展[6, 7], 有成为结肠癌预后指标的潜力。然而, 单纯免疫检查点基因的预测价值在结肠癌患者预后或免疫状态是潜力有限, 因此对免疫检查点相关基因表达谱及其预后意义的系统性分析则显得十分重要。
鉴于免疫检查点基因在结肠癌生长控制和癌症抑制中的关键作用, 我们系统地分析了肿瘤基因组图谱(The Cancer Genome Atlas, TCGA)队列中结肠癌患者差异表达的免疫检查点相关基因, 然后构建了一个与免疫检查点相关的预后模型; 外部数据集GSE143985作为验证队列以验证我们预后模型的准确性和稳健性; 我们还利用Spearman的相关分析探讨了预后模型与免疫细胞的相关性。更进一步的, 我们基于免疫检查点风险基因以及结肠癌患者临床指标构建了列线图, 为临床预后提供一定的指导依据。
从TCGA数据库(https://portal.gdc.cancer.gov/)中的TCGA-COAD队列收集488例结肠癌患者组织和42例正常组织标准化的RNA-seq数据(每千个碱基的转录每百万映射读取的片段, Fragments Per Kilobase of exon model per Million mapped fragments, FPKM)及其相应的临床信息。从高通量基因表达(Gene Expression Omnibus, GEO)数据库中的GSE143985队列收集85例结肠癌患者的RNA微阵列数据及其相应的临床信息作为外部队列验证。去除基因表达值为0的数据, 如果发现多个值, 则使用RNA表达的平均值。使用log2(表达值+1)对基因表达值进行归一化处理。此外我们还从文献上收集了47个免疫检查点相关基因[8]。
采用R语言limma包筛选TCGA队列中肿瘤样本与正常样本之间的差异表达基因。截断值设置为log2| fold change| > 0.5, 错误发现率(false discovery rate, FDR) <0.05。使用火山图和热图对结肠肿瘤样本和正常样本的基因表达值进行可视化。使用STRING数据库(https://cn.string-db.org/)对筛选出来的差异基因进行蛋白-蛋白互作网络分析。
通过最小绝对收缩和选择算子(least absolute shrinkage and selection operator, LASSO)Cox回归分析进行缩小差异基因的范围。利用LASSO Cox回归模型系数(β ), 得到回归系数的线性组合, 进而构建结肠癌患者风险评分模型。风险评分的计算公式为:
Risk score=(mRNA1× mRNA1的表达水平)+(mRNA2× mRNA2的表达水平)……+(mRNAn× mRNAn的表达水平)
根据中位风险评分, 将结肠癌患者分为高危险组和低危险组。使用R语言survival包生成风险评分的Kaplan-Meier生存曲线用以评估分组间的生存差异, 使用R语言timeROC包生成受试者工作特征曲线(receiver operating characteristic, ROC)用以评价风险评分模型的预后预测准确性。使用GSE143985队列对风险评分模型进行了外部数据验证。
使用Spearman相关性分析, 分析风险评分模型与6种免疫细胞(B细胞, CD4+ T细胞, CD8+ T细胞, 嗜中性粒细胞, 巨噬细胞, 髓系树突状细胞)的相关性, 小于0.05的P值被认为具有统计学意义。
使用单因素变量和多因素变量Cox回归分析对风险评分模型中的基因以及临床因素(年龄, 性别, 种族, TNM分期)进行预后相关性分析, 并通过R语言forestplot包绘制森林图展示每个变量的P值和95%可信区间(confidence interval, CI)。基于多因素回归分析的结果, 使用R语言rms包构建临床预测列线图来预测1年、3年、5年结肠癌患者的总生存率。列线图可视化这些重要因素的结果, 并且患者可以通过各项临床指标和风险基因相加得到“total point”来预测单个患者的预后风险。
以上所有分析方法均由R语言(v4.0.3)实现。如果没有特殊说明, 小于0.05的P值被认为具有统计学意义。
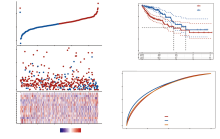
首先, 通过对比结肠癌患者和正常组织样本获得20个差异表达的免疫检查点相关基因, 其中包含7个下调基因, 13个上调基因(图1A)。热图展示了所有免疫检查点相关基因在结肠癌患者和正常组织样本的基因表达情况, 可以看到差异表达的基因在两组间有明显的颜色差异(图1B)。蛋白-蛋白互作网络分析则显示了差异表达的免疫检查点基因之间的互作关系, 在这些基因中, CD80与其他基因的互作关系最为紧密(图2A)。
使用LASSO回归分析后, 共有14个基因最终被纳入风险评分模型构建中(图2B、图2C)。其中lambda最小值为0.013 7。风险评分的计算公式如下:
Risk score=(-0.128 4)× CD160+(0.000 7)× CD200R1+(0.260 6)× CD276+(0.008 1)× CD40LG+(0.011 9)× CD44+(0.222 9)× CD80+(-0.343 1)× CTLA4+(-0.286 8)× ICOS+(0.020 8)× ICOSLG+(0.385)× LAG3+(0.19)× TNFRSF25+(0.066 6)× TNFSF15+(-0.064 8)× TNFSF9+(0.184)× VTCN1
根据中位风险评分, 将结肠癌患者分为高危险组(n=227)和低危险组(n=227)。如图3A所示, 随着风险评分的增加, 结肠癌患者的死亡人数也在显著上升, 风险基因的表达也随着风险评分的增加呈现明显的差异。Kaplan-Meier曲线表明高危组患者的总生存率明显差于低危组患者(图3B, P=1.83× 10-5)。采用随时间变化的ROC曲线评价风险评分的预后预测性能, 结果显示1年时曲线下面积(area under the curve, AUC)达到0.735, 3年时达到0.748(图3C)。
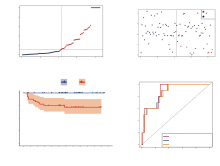
使用GEO数据库中的结肠癌患者队列GSE143985对风险评分模型进行了进一步的验证。同样的, 根据中位风险评分(图4A), 结肠癌患者被分为了高危险组(n=42)和低危险组(n=43)。如图4A所示, 随着风险评分的增加, 结肠癌患者的生存时间明显减少。其中所有死亡的结肠癌患者都具有高风险评分, 显示了风险评分精准的分层效力。Kaplan-Meier曲线表明高危组患者的总生存率明显差于低危组患者(图4B, P=3.857× 10-4)。采用随时间变化的ROC曲线评价风险评分的预后预测性能, 结果显示1年时AUC达到0.878, 3年时达到0.868, 5年时达到0.862(图4C)。
为进一步探讨风险评分与样本免疫状态的相关性, 我们使用了Spearman相关分析, 分析风险评分模型与6种免疫细胞的相关性。如图5所示, 风险评分与CD4+ T细胞呈现显著正相关(图5B, P=0.024), 与CD8+ T细胞呈现显著负相关(图5C, P=1.91× 10-4), 与其他免疫细胞相关性没有统计学差异(图5A, 图5D~F)。
单因素(图6A)回归分析和多因素(图6B)回归分析的结果显示CD40LG、CD80、ICOS、LAG3、TNFRSF25、VTCN1、年龄、TNM分期是与预后显著相关的因素。基于多因素回归分析的结果, 我们构建了结合临床指标和风险基因的预测列线图(图6C)。单个患者可以通过汇总个人具体的变量数值, 对应上每个变量对应的刻度“point”。将所有变量取值并相加合计得到总得分“total point”, 便可一次推测患者未来的1年、3年、5年的总生存率。本文中的列线图的C-指数为0.767, 显示了极佳的预测效能。其中校准曲线显示列线图模型接近对角虚线, 说明模型预测结果良好(图6D)。
结肠癌是一种预后不良的消化道恶性肿瘤, 其发病率和死亡率只增不减。近年来, 越来越多的研究致力于识别结肠癌患者中差异表达的生物标志物候选物, 以期改善患者的治疗效果和预后情况。尽管各种肿瘤标志物已被广泛应用于临床实践, 但仅靠一种生物标志物可能不足以可靠地预测患者长期结果以及预后状态。相比之下, 多基因标签可以实现更好的预测性能, 促进临床医生准确判断患者预后、区分亚群、并选择合适的治疗方法。近年来, 以PD-1/PD-L1为靶点的免疫疗法逐渐兴起, 在结肠癌患者的治疗中得到了广泛的关注[9, 10]。越来越多的证据提示免疫检查点抑制是治疗结肠癌最有希望的方法之一。然而, 值得注意的是, 当前免疫检查点抑制剂的持久的缓解率仍然很低[11]。因此, 识别引导个性化治疗的生物标志物, 有效利用医疗资源, 预防过度治疗引起的不良事件, 是目前尚未满足的需求。除PD-L1外, 其他一些免疫检查点分子包括TIGIT、TIM-3、LAG-3和CTLA-4[12, 13, 14]被提出参与结肠肿瘤微环境的免疫调节。然而, 其确切的预后价值仍有争议, 有待进一步探讨。
在本研究中, 我们通过查阅文献, 收集了47个免疫检查点基因, 并提取TCGA数据库中的基因表达谱。首先, 在正常组和结肠癌患者间筛选出29个差异表达的免疫检查点基因。随后, 利用LASSO回归最小化过拟合, 构建了一个最优的14基因预后模型。该预后模型使我们能够根据患者的风险等级将所有患者分为两个预后队列。相较于低危组患者, 高危组的生存状态明显较差。ROC曲线分析预测1年、3年的生存AUC值均大于0.7, 显示了模型较好的预测能力。同时, 上述结果也在GEO队列中得到了很好的外部验证, 其预测的1年、3年、5年的生存AUC值均大于0.8, 结果比较理想。有趣的是, 我们发现之前的研究人员也曾基于免疫相关基因建立类似的预测模型[15], 而我们的模型在测试集和验证集均显示了较高的AUC指数, 反映了其更好的预测效率。此外, 我们分析了预后模型与免疫相关性, 便于临床医生更加关注患者的免疫状态。最后, 我们还创建了一个包含免疫检查点基因和临床指标的列线图, 为临床医生的预后评估提供一定的指导价值。
我们的风险标志基因由14个基因组成, 包括CD160、CD200R1、CD276、CD40LG、CD44、CD80、CTLA4、ICOS、ICOSLG、LAG3、TNFRSF25、TNFSF15、TNFSF9、VTCN1。在TCGA队列数据中, CD160、CD200R1、CD40LG、LAG3在结肠癌患者表达中呈现明显下调, CD276、CD44、CD80、CTLA4、ICOS、ICOSLG、TNFRSF25、TNFSF15、TNFSF9、VTCN1在结肠癌患者表达则呈现明显上调。CD160是CD160/HVEM/Light/BTLA通路的重要成员, 其在免疫中的作用已被广泛认识[16], 而CD200R1表达较高的肿瘤具有“富含免疫力”的微环境[17]。CD40LG是一种CD40配体, 在T细胞表面表达, 其在结肠癌中与CD40配体的相互作用可促进T细胞依赖的B细胞增殖, 调节B细胞等类型的切换和迁移, 延长抗原特异性高亲和力记忆B细胞的存活[18]。LAG-3主要负责肿瘤细胞的免疫调节, 有研究表明在Ⅱ 期结肠癌的患者中, 肿瘤浸润性淋巴细胞上LAG-3的表达与较好的5年无进展生存期显著相关[19]。在结肠癌患者中表达明显上调的基因中, CTLA4已被证实在微卫星高度不稳定结直肠癌患者中表达升高, 这为免疫治疗在该群患者中发挥效益提供了理论基础[20]。CD276、CD44以及CD80在多种肿瘤中都有过表达, 而在结肠癌中, CD276核表达与无转移、疾病特异性和总生存率降低独立且显著相关[21], CD44表达增强了结肠癌细胞对抗癌药物(氟尿嘧啶和奥沙利铂)的耐药性[22], 而CD80表达可以促进结肠癌前病变的免疫监测[23]。ICOS和ICOSLG是诱导T细胞共刺激分子的受体和配体, 能通过共同刺激记忆T细胞功能调节二次免疫反应。有研究显示ICOS/ICOSL通路中基因的多态性可能影响晚期结肠癌患者卡培他滨化疗的疗效和不良事件的发生[24]。TNFRSF25、TNFSF15和TNFSF9均属于肿瘤坏死因子超家族成员, 主要在T细胞表达, 但其在结肠癌的作用仍未有相关报道。同样地, VTCN1已被证实在几种肿瘤中表达增加, 并有助于免疫抑制[25], 但其在结肠癌中的作用仍未见广泛报道, 这有待于进一步的研究, 并有望用作新的生物标志物。
既往研究证实, 免疫浸润是影响免疫治疗临床疗效的关键因素[26]。我们的研究结果表明, 免疫细胞浸润水平与我们构建的预后模型有显著相关性。其中风险评分与CD4+ T细胞表达水平显著正相关, 而与CD8+ T细胞表达水平显著负相关。CD4+ T细胞是初始T细胞在受到肿瘤抗原攻击后, 分化产生的一种效应T细胞, 主要起到警告和激活其他免疫细胞分子的作用[27]。在我们的研究中, 风险评分升高, CD4+ T细胞水平也显著上升, 这表明高风险人群的肿瘤抗原数量也显著提升。CD8+ T细胞科可以分化为细胞毒性细胞, 起到抵制外来病原体的入侵的作用。CD8+ T细胞浸润水平的升高可以使肿瘤对免疫治疗产生敏感反应, 其浸润水平的下降往往预示着抗肿瘤免疫水平的降低[28, 29]。这与我们的研究一致, 即高风险患者可能具有较低的抗肿瘤免疫水平。因此, 目前的研究结果可能对识别具有较高可能性从免疫治疗中获益的患者具有重要意义。
尽管如此, 我们的研究还是有一些局限性。首先, 虽然我们利用了GEO数据库中的结肠癌患者数据进行了预后模型的外部验证, 但仍然缺乏大样本的队列验证来使我们的研究具有更好的临床意义。其次, 关于免疫检查点基因在结肠癌患者中的具体生物学功能还需要体内外实验的进一步探索, 以最终为免疫检查点基因在结肠癌患者中的临床实践奠定一定的理论基础。
总之, 我们基于14个免疫检查点基因建立并验证了一个新的结肠癌预后模型, 该模型可以用于预测结肠癌患者的生存和免疫状态, 对临床预后具有一定的指导依据。
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
|